Research Focus, Project Management, Collaboration
Highlights
*
Highlights *
1) Managed 4+ research projects in Computational Chemistry, publishing findings in a 200-page technical document and delivering presentations at 5 scientific conferences.
2) Collaborated with over 3 teams of experts (including chemists, biologists, and engineers), co-authoring 5+ research articles in peer-reviewed, academic journals.
3) Designed a stand-alone medicinal chemistry project to study disease mechanisms using computational approaches, by reviewing 30+ research papers across 10+ databases; authored a detailed report and presented findings to leadership.
4) Developed scientific expertise in the following areas
-
Structural Biology, Biophysics, Biochemistry, Mechanobiology
-
Statistical Mechanics, Chemical Physics, Physical Chemistry
-
Statistics, Machine Learning (ML), Artificial Intelligence (AI)
Below is a list of some of my projects and research articles
2024
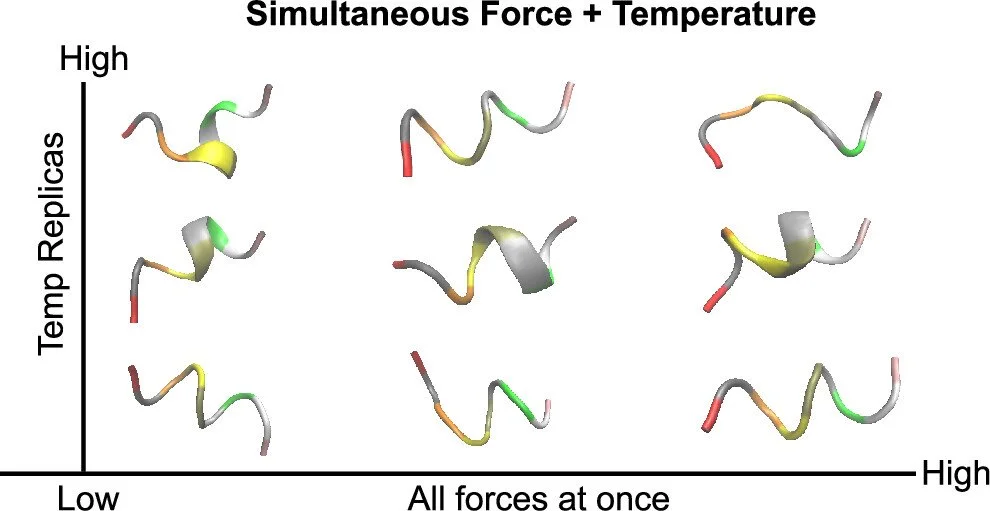
Improved Prediction of Molecular Response to Pulling by Combining Force Tempering with Replica Exchange Methods
In cell processes, tiny forces are crucial, like in the movement of cell structures and controlling pressure in cells. Computer simulations can help design models that respond to these forces, but it's hard to predict their effects because they are so small. In this project, I show that the combination of multiple computational research techniques is not only sound in theory but is also a robust approach to collect data 8 times faster. Code, data, and tutorials for this project is publicly available, free of charge here.
2023
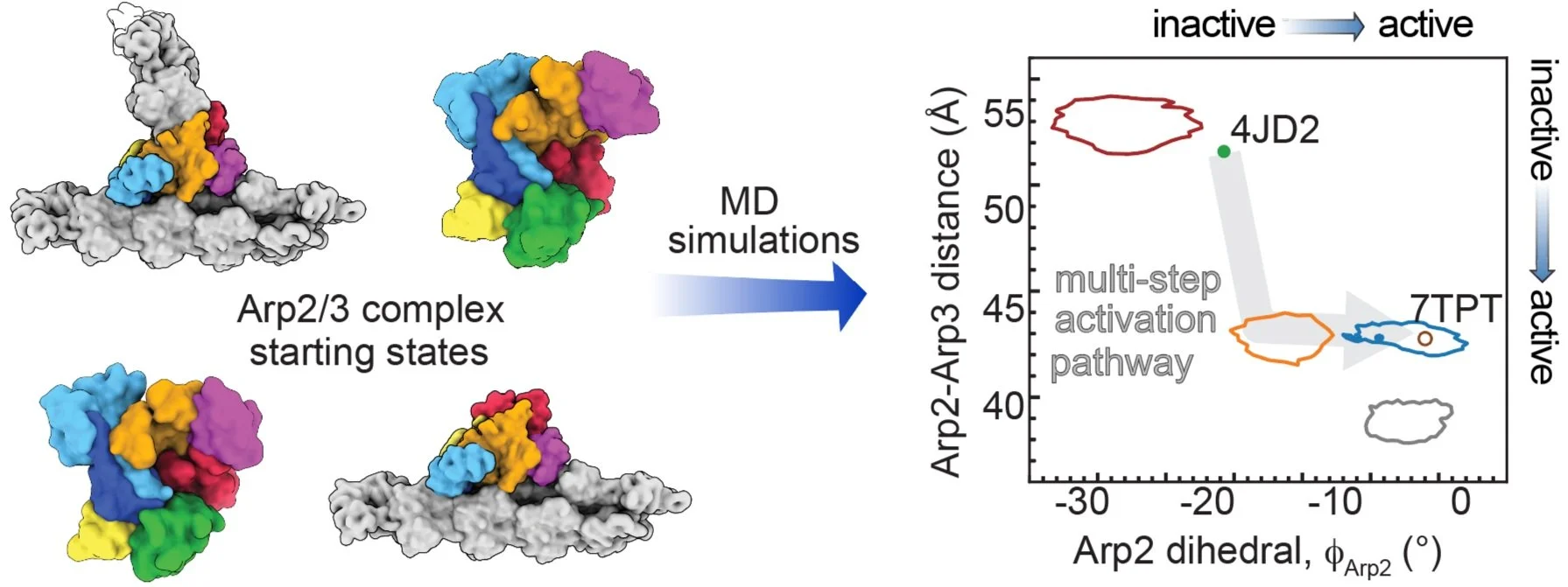
Molecular dynamics simulations support a multistep pathway for activation of branched actin filament nucleation by Arp2/3 complex
The Arp2/3 complex is like a molecular machine that helps create branch-like structures in cells. It's crucial for processes like cell movement. When activated by certain proteins, it undergoes changes in its shape. In this project, I constructed over 4 computational models and generated up to 10GB of numerical data that agrees with experimental findings by over 80%. Code, data, and tutorials for this project is publicly available, free of charge here.
2022
Structure of Arp2/3 complex at a branched actin filament junction resolved by single-particle cryo-electron microscopy
Imagine the Arp2/3 complex as a microscopic powerhouse inside cells, teaming up with other molecular machinery to orchestrate vital cellular functions, such as growth and movement. Despite years of investigation, no one completely understands the mysteries of this machinery and how cells manage it. In a joint effort, I collaborated with biochemists to develop a computational model. This model generated numerical data remarkably close, within a 2% margin of error, to results obtained through sophisticated biochemistry experiments.
2021
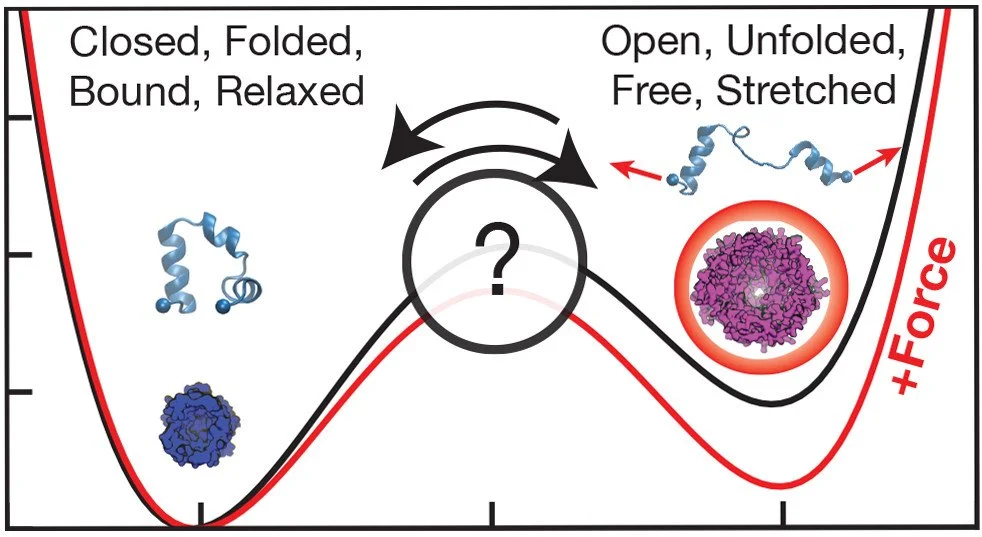
Molecular Paradigms for Biological Mechanosensing
Within cells, proteins frequently encounter minuscule forces originating from internal sources like molecular machines or external factors such as pressure. Despite their small scale, specific proteins, known as "mechanosensing" proteins, possess the ability to sense and respond to these forces. In this perspective article, I played a role in parsing through over 10 previous scientific paper and summarizing 1 of the 5 types of proteins discussed in this context.
2020
Infinite switch simulated tempering in force (FISST)
Proteins in cells can feel tiny forces, and even small changes matter for their jobs. To study these forces, I contributed to a method that helps explore how proteins react when we pull on them with different strengths. Testing with 5 different mechanical forces on 3 computational models gives performance of at least 90% accuracy. The code and data are available publicly, free of charge here.